Nearly 1,000 Days Late, US Senate Agrees with Me and Concludes Laboratory Spread Most Likely. Plus: Contrary to What Chinese Scientists Said, pShuttle-SN is closer to SARS-CoV-2 than to SARS-CoV-1
As the first (or at least one of the first rare) scientist to report that it likely came from the lab, the US Senate is now in agreement using a consilience of the evidence. About pShuttle...
On January 30, 2020, I published this article:
That makes me if not among the first, then by far, among the first, to propose a lab origin of the SARS-CoV-2 virus. It created quite a storm.
This month, October, 2022, US Senate:
The reasons why the US Senate finally agree:
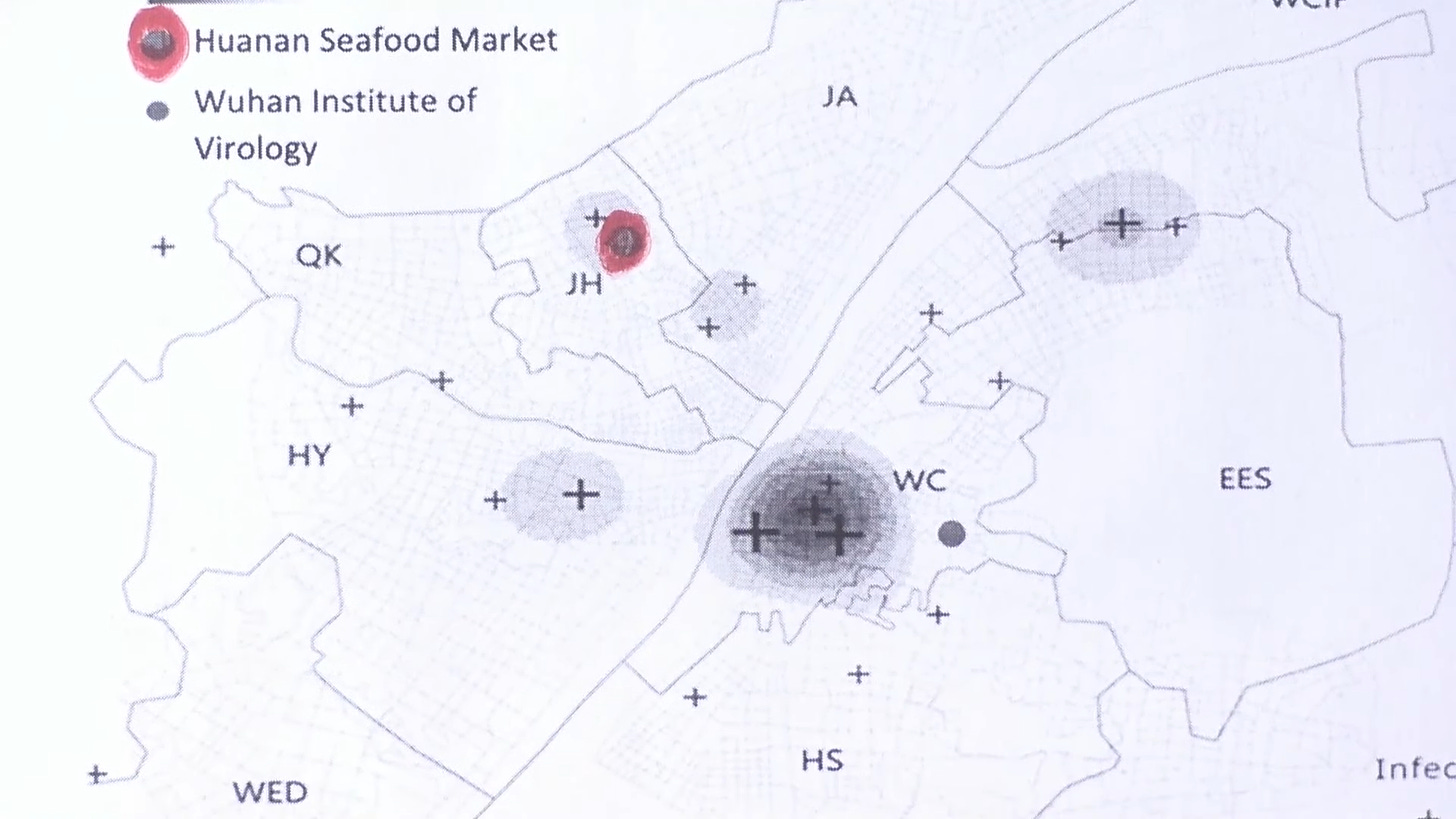
The internet searches on flu-like symptoms shows the epicenter was nearthe Wuhan Institute for Virology, not the Hunan Market.
From John Campbell’s excellent review of this report:
The virus was too well-adapted to human-to-human transmission.
No convincing intermediate host has been found.
Past outbreaks of respiratory viruses involving “spillovers” took longer, and involved more transmission events.
Low genetic variation among the among variants of SARS-CoV-2 (only 2 nucleotide differences) indicate single source of a ready-made virus.
Multiple laboratory leaks of SARS-CoV-1. Ironically, these had led to the ban on Gain-of-Function Research violated by NIAID’s act of funding WIV to continue to conduct such research.
Before we get to the pShuttle-N business, here’s John’s summary.
Back in early 2020, I had considered the specific link between the “missing fragment” that somehow matched a sequence in a pShuttle-SN.
By March, 2020, the CCP had put these scientists to the task of rebutting my hypotheses:
Is SARS-CoV-2 originated from laboratory? A rebuttal to the claim of formation via laboratory recombination
Pei Hao, Wu Zhong Shiyang Song Shiyong Fan and Xuan Li
There’s my name, the first line of a peer-reviewed article even, received Feb 19, 2020.
These scientists wrote the “Plasmid generated from Adeno-X(TM) to study SARS-CoV”, not pShuttle-SN, and that somehow I was confused by saying the pShuttle-SN was a vector.
They also wrote:
“On an added note, our results indicated its sequence similarity to the pShuttle-SN fragment (Supplementary Figure 2) was lower than to the natural coronaviral sequences (Figure 1).”
The CCP censors did not catch the tell.
Can you see it? (Drop a comment if you see it in their text).
Go to NCBI, and you find that pShuttle-SN is called
"Expression vector pShuttle-SN, complete sequence" https://www.ncbi.nlm.nih.gov/nuccore/AY862402
Why would Chinese scientists try to convince the world that an expression vector was not be called a vector?
Notably, they referenced my second, not my first article.
Out of curiosity, yesterday, I decided to Blast the Amino Acid Sequenced predicted by NCBI’s ORF finder from the “inserted portion” I had reported as found.
Guess what?
This came at 100% sequence similarity at the protein level:
Range 1: 153 to 504GenPeptGraphicsNext MatchPrevious Match
Alignment statistics for match #1ScoreExpectMethodIdentitiesPositivesGaps735 bits(1897)0.0Compositional matrix adjust.352/352(100%)352/352(100%)0/352(0%)
Query 1 MESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINL 60
MESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINL
Sbjct 153 MESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINL 212
Query 61 VRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQP 120
VRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQP
Sbjct 213 VRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQP 272
Query 121 RTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNI 180
RTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNI
Sbjct 273 RTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNI 332
Query 181 TNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCF 240
TNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCF
Sbjct 333 TNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCF 392
Query 241 TNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYL 300
TNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYL
Sbjct 393 TNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYL 452
Query 301 YRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVG 352
YRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVG
Sbjct 453 YRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVG 504Look how nice a clean that alignment is!
Now look at the next best match, SARS-CoV-1 (63%)
RecName: Full=Spike glycoprotein; Short=S glycoprotein; AltName: Full=E2; AltName: Full=Peplomer protein; Contains: RecName: Full=Spike protein S1; Contains: RecName: Full=Spike protein S2; Contains: RecName: Full=Spike protein S2'; Flags: Precursor [Severe acute respiratory syndrome-related coronavirus]
Sequence ID: P59594.1Length: 1255Number of Matches: 1
Range 1: 147 to 490GenPeptGraphicsNext MatchPrevious Match
Alignment statistics for match #1ScoreExpectMethodIdentitiesPositivesGaps471 bits(1212)7e-157Compositional matrix adjust.221/351(63%)270/351(76%)7/351(1%)
Query 2 ESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLV 61
++ ++ +A NCTFEY+S F +D+ K GNFK+LREFVFKN DG+ +Y + PI++V
Sbjct 147 QTHTMIFDNAFNCTFEYISDAFSLDVSEKSGNFKHLREFVFKNKDGFLYVYKGYQPIDVV 206
Query 62 RDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPR 121
RDLP GF+ L+P+ LP+GINIT F+ +L + +P W AAAY+VGYL+P
Sbjct 207 RDLPSGFNTLKPIFKLPLGINITNFRAIL----TAFSPAQDI--WGTSAAAYFVGYLKPT 260
Query 122 TFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNIT 181
TF+LKY+ENGTITDAVDC+ +PL+E KC++KSF ++KGIYQTSNFRV P+ +VRFPNIT
Sbjct 261 TFMLKYDENGTITDAVDCSQNPLAELKCSVKSFEIDKGIYQTSNFRVVPSGDVVRFPNIT 320
Query 182 NLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFT 241
NLCPFGEVFNAT+F SVYAW RK+ISNCVADYSVLYNS FSTFKCYGVS TKLNDLCF+
Sbjct 321 NLCPFGEVFNATKFPSVYAWERKKISNCVADYSVLYNSTFFSTFKCYGVSATKLNDLCFS 380
Query 242 NVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLY 301
NVYADSFV++GD+VRQIAPGQTG IADYNYKLPDDF GCV+AWN+ N+D+ GNYNY Y
Sbjct 381 NVYADSFVVKGDDVRQIAPGQTGVIADYNYKLPDDFMGCVLAWNTRNIDATSTGNYNYKY 440
Query 302 RLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVG 352
R R L+PFERDIS + PC NCY+PL YGF T G+G
Sbjct 441 RYLRHGKLRPFERDISNVPFSPDGKPCTP-PALNCYWPLNDYGFYTTTGIG 490
Not so clean, eh?
The next one? HKU3, which I told everyone needed to be focused on as the likely backbone:
RecName: Full=Spike glycoprotein; Short=S glycoprotein; AltName: Full=E2; AltName: Full=Peplomer protein; Contains: RecName: Full=Spike protein S1; Contains: RecName: Full=Spike protein S2; Contains: RecName: Full=Spike protein S2'; Flags: Precursor [Bat SARS coronavirus HKU3]Sequence ID: Q3LZX1.1Length: 1242Number of Matches: 1
Range 1: 156 to 472GenPeptGraphicsNext MatchPrevious Match
Alignment statistics for match #1ScoreExpectMethodIdentitiesPositivesGaps409 bits(1050)3e-133Compositional matrix adjust.200/341(59%)254/341(74%)24/341(7%)
Query 7 VYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQ 66
VY SA NCT++ V + F +D K GNFK+LRE+VFKN DG+ +Y +T +NL R LP
Sbjct 156 VYQSAFNCTYDRVEKSFQLDTTPKTGNFKDLREYVFKNRDGFLSVYQTYTAVNLPRGLPT 215
Query 67 GFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLK 126
GFS L+P++ LP GINIT ++ ++A+ ++S + +AAYYVG L+ TF+L+
Sbjct 216 GFSVLKPILKLPFGINITSYRVVMAMF------SQTTSNFLPESAAYYVGNLKYSTFMLR 269
Query 127 YNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPF 186
+NENGTITDAVDC+ +PL+E KCT+K+F V+KGIYQTSNFRV PT+ ++RFPNITN CPF
Sbjct 270 FNENGTITDAVDCSQNPLAELKCTIKNFNVDKGIYQTSNFRVSPTQEVIRFPNITNRCPF 329
Query 187 GEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYAD 246
+VFNATRF +VYAW R +IS+CVADY+VLYNS SFSTFKCYGVSP+KL DLCFT+VYAD
Sbjct 330 DKVFNATRFPNVYAWERTKISDCVADYTVLYNSTSFSTFKCYGVSPSKLIDLCFTSVYAD 389
Query 247 SFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRK 306
+F+IR EVRQ+APG+TG IADYNYKLPDDFTGCVIAWN+ D+ NY YR RK
Sbjct 390 TFLIRSSEVRQVAPGETGVIADYNYKLPDDFTGCVIAWNTAKHDTG-----NYYYRSHRK 444
Query 307 SNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQP 347
+ LKPFERD+S++ NGV + L +Y F P
Sbjct 445 TKLKPFERDLSSD-------DGNGV------YTLSTYDFNP 472What About RATg13?
Hao et al., published that similarity to RATg13 was higher… so curious me… found 94% similarity in these amino acid positions to the pShuttle-SN expression vector.
Now, RatG13 has an interesting history. The genome sequence was published 3/20/2020… after my article.
Further, Dr. Shi reported to the NY Times that RatG13 could not be the backbone used (96% similarity is not close enough)…
Related: See RedState’s Wuhan's 'Bat Woman' Straight-Up Lied to the New York Times About Her Research
Here’s the RatG13 result; only 94% across the spike protein.
lcl|ORF1:323:1378 unnamed protein product, partialSequence ID: Query_43489Length: 352Number of Matches: 2
Range 1: 1 to 352GraphicsNext MatchPrevious Match
Alignment statistics for match #1ScoreExpectMethodIdentitiesPositivesGaps694 bits(1791)0.0Compositional matrix adjust.331/352(94%)339/352(96%)0/352(0%)
Query 153 MESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINL 212
MESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINL
Sbjct 1 MESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINL 60
Query 213 VRDLPPGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQP 272
VRDLP GFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQP
Sbjct 61 VRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQP 120
Query 273 RTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTDSIVRFPNI 332
RTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPT+SIVRFPNI
Sbjct 121 RTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNI 180
Query 333 TNLCPFGEVFNATTFASVYAWNRKRISNCVADYSVLYNSTSFSTFKCYGVSPTKLNDLCF 392
TNLCPFGEVFNAT FASVYAWNRKRISNCVADYSVLYNS SFSTFKCYGVSPTKLNDLCF
Sbjct 181 TNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCF 240
Query 393 TNVYADSFVITGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSKHIDAKEGGNFNYL 452
TNVYADSFVI GDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNS ++D+K GGN+NYL
Sbjct 241 TNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYL 300
Query 453 YRLFRKANLKPFERDISTEIYQAGSKPCNGQTGLNCYYPLYRYGFYPTDGVG 504
YRLFRK+NLKPFERDISTEIYQAGS PCNG G NCY+PL YGF PT+GVG
Sbjct 301 YRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVG 352I make mistakes, and I’m the first to admit and correct them when I can. I am honest, because science demands integrity first and have no desire or reason to mislead.
I am calling on Wikipedia and Twitter to reverse course and get back in touch with reality.
I won’t put the Chinese scientists in a bad position with the CCP by demanding retraction, but instead I wish them well and safe passage in their journey.






the pShuttle-N was "built" in 2005?
I believe the tell is the word natural.
Thanks for your articles and insights. You always entertain and inform. Thank you!